publications
publications by categories in reversed chronological order. generated by jekyll-scholar.
2025
- J Comput Neurosci
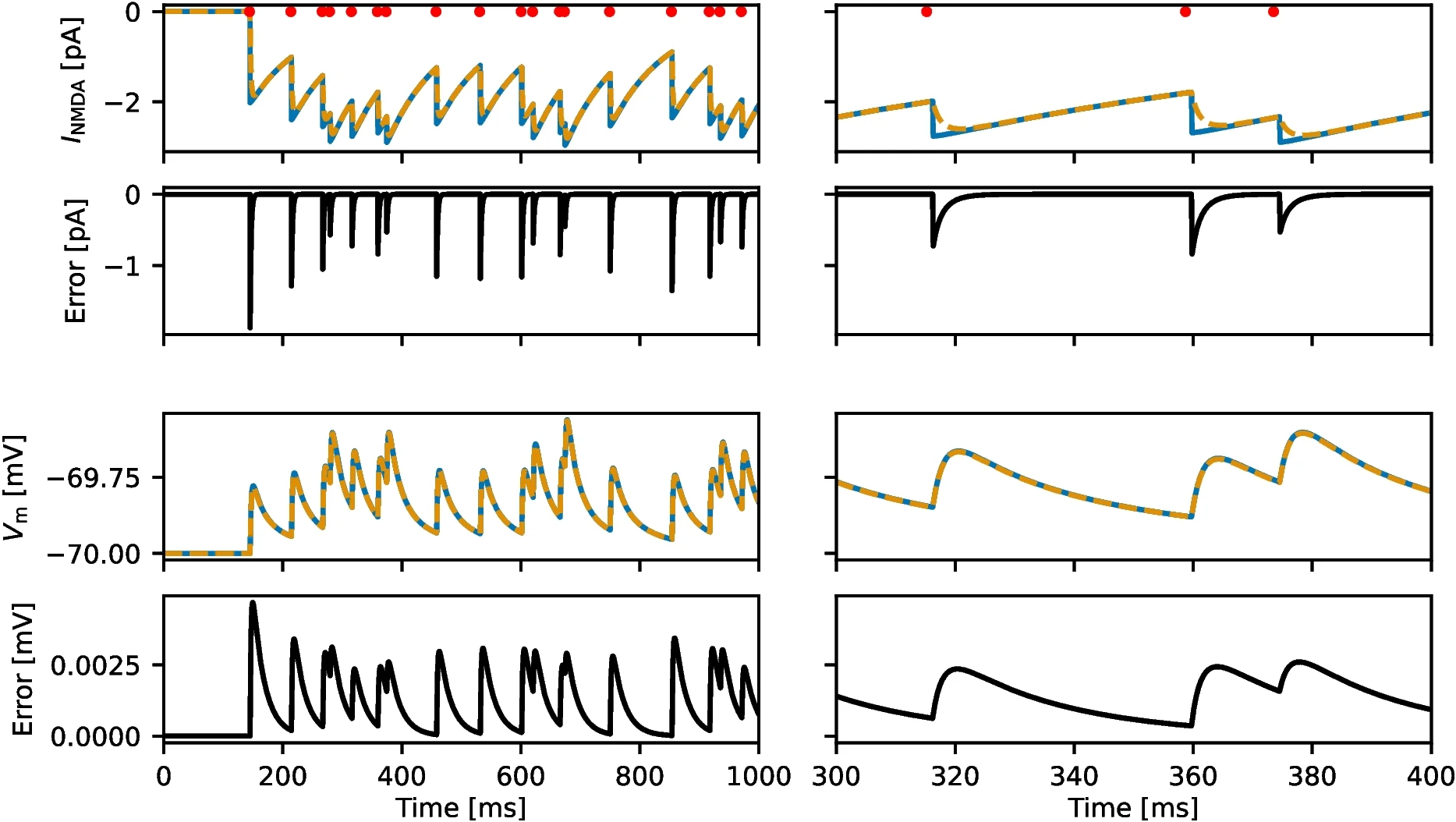 A simplified model of NMDA-receptor-mediated dynamics in leaky integrate-and-fire neuronsJan-Eirik W. Skaar, Nicolai Haug, and Hans Ekkehard PlesserJournal of Computational Neuroscience, Aug 2025
A simplified model of NMDA-receptor-mediated dynamics in leaky integrate-and-fire neuronsJan-Eirik W. Skaar, Nicolai Haug, and Hans Ekkehard PlesserJournal of Computational Neuroscience, Aug 2025A model for NMDA-receptor-mediated synaptic currents in leaky integrate-and-fire neurons, first proposed by Wang (J Neurosci, 1999), has been widely studied in computational neuroscience. The model features a fast rise in the NMDA conductance upon spikes in a pre-synaptic neuron followed by a slow decay. In a general implementation of this model which allows for arbitrary network connectivity and delay distributions, the summed NMDA current from all neurons in a pre-synaptic population cannot be simulated in aggregated form. Simulating each synapse separately is prohibitively slow for all but small networks, which has largely limited the use of the model to fully connected networks with identical delays, for which an efficient simulation scheme exists. We propose an approximation to the original model that can be efficiently simulated for arbitrary network connectivity and delay distributions. Our results demonstrate that the approximation incurs minimal error and preserves network dynamics. We further use the approximate model to explore binary decision making in sparsely coupled networks.
@article{10.1007/s10827-025-00911-8, doi = {10.1007/s10827-025-00911-8}, author = {Skaar, Jan-Eirik W. and Haug, Nicolai and Plesser, Hans Ekkehard}, journal = {Journal of Computational Neuroscience}, publisher = {Springer Nature}, title = {A simplified model of NMDA-receptor-mediated dynamics in leaky integrate-and-fire neurons}, year = {2025}, month = aug, url = {https://doi.org/10.1007/s10827-025-00911-8}, }
2023
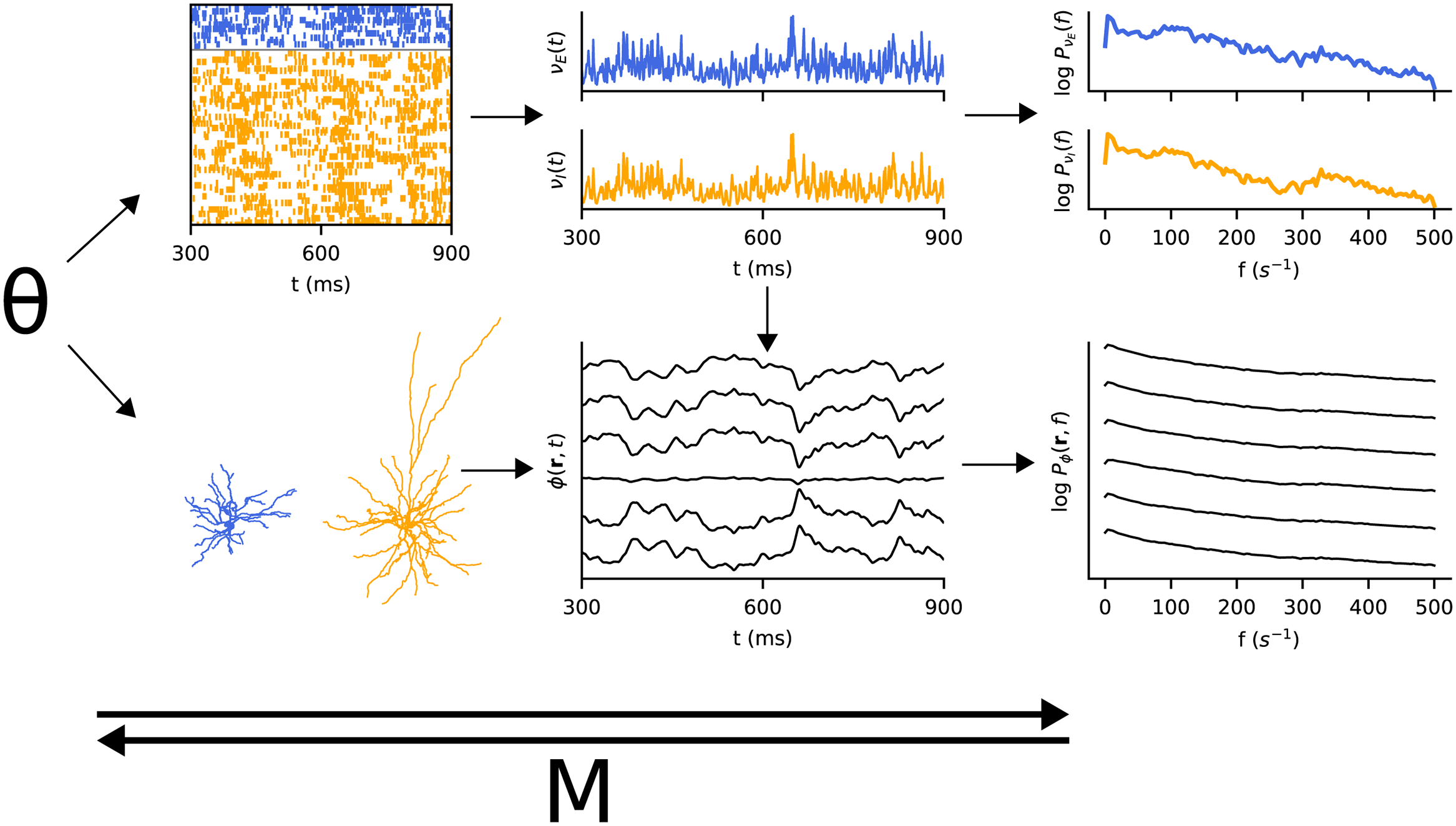
- PLoS Comput. BiolMetamodelling of a two-population spiking neural networkJan-Eirik W. Skaar, Nicolai Haug, Alexander J. Stasik, and 2 more authorsPLOS Computational Biology, Nov 2023
In computational neuroscience, hypotheses are often formulated as bottom-up mechanistic models of the systems in question, consisting of differential equations that can be numerically integrated forward in time. Candidate models can then be validated by comparison against experimental data. The model outputs of neural network models depend on both neuron parameters, connectivity parameters and other model inputs. Successful model fitting requires sufficient exploration of the model parameter space, which can be computationally demanding. Additionally, identifying degeneracy in the parameters, i.e. different combinations of parameter values that produce similar outputs, is of interest, as they define the subset of parameter values consistent with the data. In this computational study, we apply metamodels to a two-population recurrent spiking network of point-neurons, the so-called Brunel network. Metamodels are data-driven approximations to more complex models with more desirable computational properties, which can be run considerably faster than the original model. Specifically, we apply and compare two different metamodelling techniques, masked autoregressive flows (MAF) and deep Gaussian process regression (DGPR), to estimate the power spectra of two different signals; the population spiking activities and the local field potential. We find that the metamodels are able to accurately model the power spectra in the asynchronous irregular regime, and that the DGPR metamodel provides a more accurate representation of the simulator compared to the MAF metamodel. Using the metamodels, we estimate the posterior probability distributions over parameters given observed simulator outputs separately for both LFP and population spiking activities. We find that these distributions correctly identify parameter combinations that give similar model outputs, and that some parameters are significantly more constrained by observing the LFP than by observing the population spiking activities.
@article{10.1371/journal.pcbi.1011625, doi = {10.1371/journal.pcbi.1011625}, author = {Skaar, Jan-Eirik W. and Haug, Nicolai and Stasik, Alexander J. and Einevoll, Gaute T. and Tøndel, Kristin}, journal = {PLOS Computational Biology}, publisher = {Public Library of Science}, title = {Metamodelling of a two-population spiking neural network}, year = {2023}, month = nov, volume = {19}, url = {https://doi.org/10.1371/journal.pcbi.1011625}, pages = {1-26}, number = {11}, }